18. Troubleshooting¶
18.1. Installation¶
18.1.1. You don’t know where to put the bamm directory¶
On OS X, you may move the uncompressed bamm directory (folder) to your home
directory. For example, if it was uncompressed in your Downloads directory:
mv ~/Downloads/bamm-1.0.0 ~
To run BAMM within your data directory, you will need to specify its full location (path):
~/bamm-1.0.0/bamm -c divcontrol.txt
On Windows, you may move the uncompressed “bamm” directory (folder) to your
C: drive. To run BAMM within your data directory, you will need to specify
its full path:
c:\bamm\bamm -c divcontrol.txt
18.2. Running¶
18.2.1. On OS X, you get an error that says something like “dyld: unknown required load command 0x80000022”¶
This likely means that you are trying to run BAMM on an older version of OS X. Please run BAMM on OS X 10.5 or greater.
18.2.2. On Windows, you get an error that says “bamm.exe is not a valid Win32 application”¶
This likely means that you are trying to run BAMM on an unsupported version of Windows (such as Windows XP). Please run BAMM on Windows 7 or greater.
18.2.3. On Unix/Linux, you get the error message “Enable multithreading to use std::thread: Operation not permitted”¶
We have found this problem with some versions of the GNU C++ compiler, where CMake does not properly link a required threading library. We are working on a solution, but in the meantime, you can manually compile BAMM (without using CMake) as follows:
g++ -DBAMM_VERSION=\"2.2.0\" -DBAMM_VERSION_DATE=\"2014-09-05\" -DGIT_COMMIT_ID=\"NA\" -pthread -std=c++11 -o bamm ../src/\*.cpp
Make sure to run this command in an empty build directory.
18.2.4. You get an error message by BAMM¶
Most error messages in BAMM are descriptive and therefore it should not be
difficult to correct the problem. If BAMM cannot find the control file or
any file specified within the control file, such as the tree file, make sure
that the file name is spelled correctly and that the location given in the
control file is relative to the directory in which bamm was called.
Some error messages in BAMM are more cryptic and may require that you contact
us directly.
Please go to Contact Us on
instructions on how to contact us.
18.2.5. Basic troubleshooting¶
The simplest way to localize potential problems is to first try to load your data without doing anything else. You can do this by setting the following parameter in your controlfile:
initializeModel = 0
This tells BAMM to simply try reading the tree (and associated trait or taxon sampling datafiles). If this works fine, your problem is probably elsewhere. The next step is to try initializing your MCMC simulation, without actually running it. This step involves actually computing likelihoods and priors, so may reveal problems with those steps:
initializeModel = 1
runMCMC = 0
If these issues fail to resolve the problem, please contact us. Go to Contact Us on instructions on how to contact us.
18.2.6. You get an operating system error (e.g., segmentation fault)¶
Other possible errors may arise, not discussed above. This may include errors that mention “Segmentation fault”, or “Floating point error”. Please contact us with information about system errors. Go to Contact Us on instructions on how to contact us.
18.3. Analysis¶
18.3.1. The MCMC does not converge¶
Achieving satisfactory convergence with BAMM can be difficult. We know that the likelihood surface for the BAMM model is rugged, with lots of peaks and valleys. We are developing several new MCMC proposals for moving, splitting, and merging events on trees that should facilitate convergence. In the meantime, we suggest several options.
First, don’t be afraid to run BAMM for a really, really long time if you are having convergence issues. The implementation is fast. We suggest sampling very infrequently so as to not generate overwhelming quantities of output. Running BAMM for 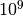 generations, sampling parameters every
generations, sampling parameters every 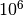 generations, is not unreasonable for some datasets.
generations, is not unreasonable for some datasets.
Second, make sure your priors are not maladjusted. For example, if you have a phylogenetic tree that is calibrated in units of substitutions (rather than millions of years), the default priors in BAMM’s control file could lead to significant difficulties. We suggest using the BAMMtools function setBAMMpriors to choose priors that are reasonably matched to your dataset. There is no guarantee that these priors will be any good, but they are at least reasonably scaled to your data.
Third: one of the most common problems in moving between “peaks” on the macroevolutionary rate shift landscape is that peaks are separated by valleys that require multiple moves to traverse. You might have an optimum at N = 7 shifts, and another at N = 3 shifts, and both of these scenarios might have equivalent posterior probabilities. However, if shift configurations with 4, 5, and 6 shifts have lower posterior probabilities than either 3 or 7, it will be very hard to move between these submodels. One solution we have found to work involves increasing the value of the hyperprior on the Poisson rate parameter governing the number of rate shifts. For large datasets, we have experienced dramatic improvements in performance by flattening this distribution. To flatten this (thus allowing the algorithm to explore a broader range of shift configurations), you would choose a lower value for the poissonRatePrior. The default is 1.0, but setting this to poissonRatePrior = 0.1 or even poissonRatePrior = 0.02 (for very large trees) is reasonable and should facilitate convergence.
Fourth, try restricting the regions of the tree where shifts are permitted to occur. BAMM has an argument minCladeSizeForShift that enables you to constrain your analysis to only allow rate shifts to occur on branches that have at least minCladeSizeForShift descendants. The default value, 1, allows shifts to occur on all branches. However, if you set minCladeSizeForShift = 10, you would only allow rate shifts to occur on branches with at least 10 descendants. Thus, BAMM would not waste time putting rate shifts on terminal branches (or, for that matter, any branches with < 10 tip descendants).
Fifth, consider reducing model complexity. If you are having trouble getting convergence with the full BAMM analysis, try running your analysis after “turning off” the time-varying rates component of BAMM. Thus, you would just be running a version of BAMM that allowed mixtures of different constant-rate processes across a phylogeny. Parameter settings to run this model are explained here.
Finally: if you have tried some or all of the above, and still have issues with convergence, we encourage you to explore your data further to assess whether you should expect the BAMM model to fit your data. One recent example we have seen involved a dataset where maximum phenotypic divergences were observed between the most closely related species (suggesting the possibility of character displacement). This pattern is not something that BAMM can really model at present: even though BAMM allows phenotypic rates to vary through time, the model still predicts that, on average, phenotypic similarity should be positively related to phylogenetic relatedness.
